The choice of the classification algorithm is particularly important, as this will determine the ‘real’ classification accuracy and will ensure that the number of variables is kept to a minimum.
It is vital that the algorithm can cross-validate its output. The goal of cross-validation is to use part of the data to train the model (identify the model coefficients) and part of the data to test the model (validation process). Cross-validation allows limiting problems like overfitting and helps building a model that will have a high classification accuracy not only for the respondents in the sample used for the segmentation, but also for new respondents not included in the segmentation study.
An algorithm not tested via cross-validation might have a high prediction accuracy on the sample data (e.g., 90%), but is likely to have a much lower prediction accuracy on new data (e.g., 40%).
The only way to ensure high prediction accuracy on new data is to test the model results via cross-validation.
R-sw Discriminant is a R-sw module specifically developed for segmentation typing tools (also known as allocation tools). It is the only commercial software that:
- allows cross-validating the results during the selection process (leading to better models), not only cross-validating the discriminant model after this has been chosen;
- produces a full step-by-step report at each step of the selection process;
- has got a built-in Genetic Algorithm allowing a very efficient selection process, typically leading to the identification of a set of variables with a higher classification accuracy than standard algorithms;
- comes with a fully-customizable Excel allocation tool, allowing the user to quickly develop new accurate, user-friendly and graphically appealing allocation tools.
Example of Report (first step only)
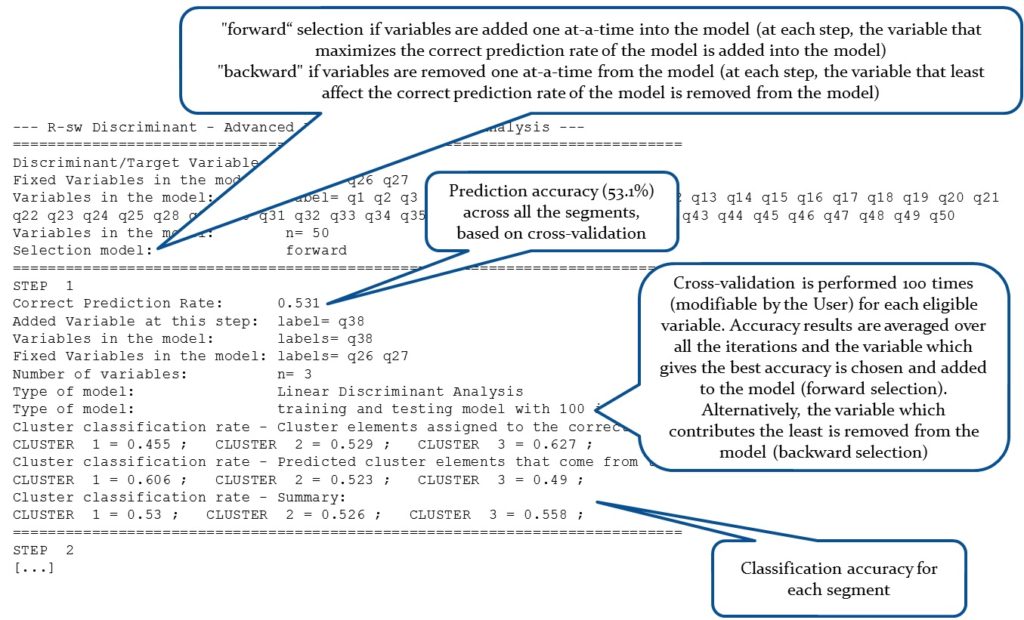
The function DA.report available in R-sw Discriminant produces a step-by-step report of the selection process, inclusive of the classification accuracy achievable by considering subsets of the specified variables. Variables are added or removed one at-a-time. A subset of variables can be forced into the models. Cross-validation through repeated random sub-sampling validation can be performed at each step to ensure minimal overfitting and thus identify relationships in the data that hold in general.
Through a Genetic Algorithm, the function genetic.DA allows the identification of the combination of variables with the highest classification accuracy from the set of variables specified. Cross-validation through repeated random sub-sampling validation can be performed at each iteration to estimate the correct prediction rate of the discriminant model.
The function discriminant.analysis performs linear or quadratic discriminant analysis on a specific set of variables and returns a set of coefficients to be used in a typing / allocation tool. Cross-validation through repeated random sub-sampling validation can be performed to estimate the correct prediction rate of the discriminant model.
The function predict.DA predicts group / cluster membership for a new set of records based on a linear or quadratic discriminant model.
